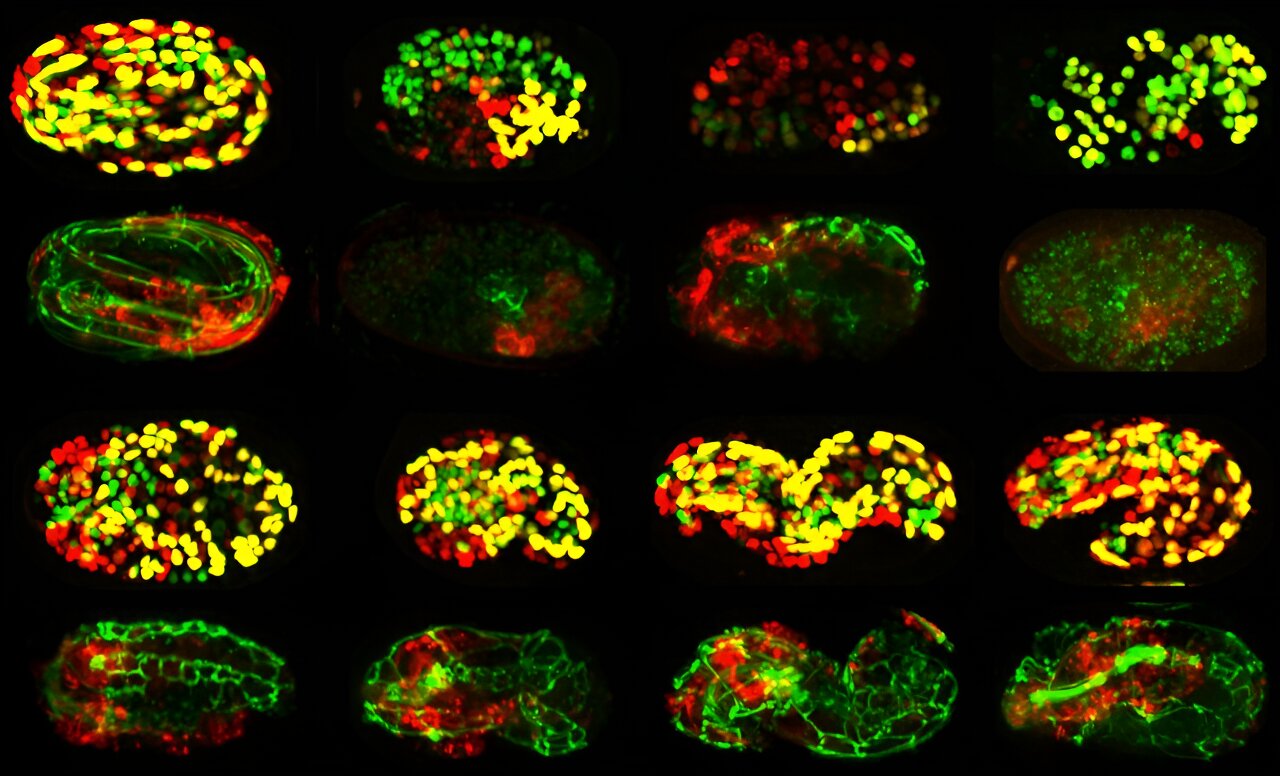
This gallery shows a collection of embryos after genes were blocked one by one. The different outcomes (or observed characteristics) for each embryo reflect the specific functions of the genes tested. Credit: Rebecca Green, Oegema Lab, UC San Diego
× close to
This gallery shows a collection of embryos after genes were blocked one by one. The different outcomes (or observed characteristics) for each embryo reflect the specific functions of the genes tested. Credit: Rebecca Green, Oegema Lab, UC San Diego
Although the Human Genome Project announced the completed sequencing of 20,000 human genes more than two decades ago, scientists are still trying to understand how fully formed beings emerge from basic genetic instructions.
Biomedical efforts to learn how disorders can manifest at the earliest stages of development would benefit from knowing specifically how complex organisms arise from a single fertilized cell. Researchers at the University of California San Diego have gained new insight into how embryonic development unfolds through the lens of a simple model organism.
The comprehensive report led by School of Biological Sciences scientist Rebecca Green and Professor Karen Oegema provides a play-by-play look at how genes function during embryonic development in Caenorhabditis elegans (C. elegans), a millimeter-long roundworm known to biologists as “the worm.” Despite its small size, C. elegans is a workhorse for scientists because much of its biology, including early stages of development, resembles that of higher organisms, including humans.
The research, which combines ten years of work by a collaborative multidisciplinary team into a ‘genetic atlas’, is published in the journal Cell.
“By characterizing many of these poorly understood genes in a simple model organism, we can learn what they do in more complex systems like humans,” says Green, a bioinformatics scientist and first author of the paper. “Although the work is done using C. elegans, the majority of the genes analyzed are present in humans and mutations in many of them are associated with human developmental disorders.”
The researchers developed an automated system for profiling the function of genes required for embryogenesis, the process by which a fertilized egg, which starts out as a single cell, develops into an organism with different tissues, such as skin, digestive tract, neurons and muscles. They used time-lapse 4D imaging to methodically track the function of each gene at all embryonic stages, including when cell identity is determined and when tissues in the organism take shape.
The researchers monitored this process using an approach known as ‘computer vision’ to monitor specific aspects of development, including the number of cells in each tissue. They also tracked the mass, position and shape of the tissues in the developing organism.
To fully understand the function of nearly 500 genes important in embryonic development, they blocked the function of each gene one by one. This allowed the researchers to group genes into common clusters that revealed each gene’s role through ‘guilt by association’. Green compares the process to automated facial recognition, which groups together images with similar features.
By using this painstaking process to analyze a collection of nearly 7,000 4D embryogenesis films, the team was able to create “fingerprints” of individual genes, such as those that cells need to become muscle or skin. This helped them understand the physiological role the genes play in embryogenesis, such as controlling the formation of tissues such as the intestines or the nervous system.
“We show that our approach correctly classifies the functions of previously characterized genes, identifies functions for poorly characterized genes, and describes novel gene and signaling pathway relationships,” said Oegema, faculty member in the Department of Cell and Developmental Biology and senior author of the paper. “Many genes that we thought performed everyday functions turned out to play important roles that were underappreciated.”
In combination with the Cell On paper, the abundance of data from the survey has led to the launch of a new online resource containing all the information. PhenoBank now provides a portal to the genetic atlas developed during the research.
The research team included (from left) Zhiling Zhao, Rebecca Green, Renat Khaliullin and Stacy Ochoa. Credit: Rebecca Green, Oegema Lab, UC San Diego
× close to
The research team included (from left) Zhiling Zhao, Rebecca Green, Renat Khaliullin and Stacy Ochoa. Credit: Rebecca Green, Oegema Lab, UC San Diego
“The approach provided surprising insights into how metabolic pathways are specialized during embryogenesis and revealed interesting new connections between different molecular machineries involved in gene regulation,” says Professor Arshad Desai, co-author of the paper.
In addition to the 500 genes contained in the Cell study, researchers are now working to complete the full sequence of 2,000 C. elegans genes involved in embryogenesis.
“The broad interest lies in the approach developed to tackle perhaps the most challenging problem in biology: how a single cell with a genome containing approximately 20,000 genes (similar to the number of genes in humans) is able to whole organism to build,” he said.
Authors of the article include Rebecca Green, Renat Khaliullin, Zhiling Zhao, Stacy Ochoa, Jeffrey Hendel, Tiffany-Lynn Chow, HongKee Moon, Ronald Biggs, Arshad Desai and Karen Oegema. The researchers also thank Tony Hyman and the Scientific Computing group of the Max Planck Institute for Molecular Cell Biology and Genetics (MPI-CBG) for facilitating the construction of PhenoBank.
More information:
Rebecca A. Green et al., Automated profiling of gene function during embryonic development, Cell (2024). DOI: 10.1016/j.cell.2024.04.012
Magazine information:
Cell